Responsible for the construction of all living things, DNA formats organic data within an organism. DNA is, in essence, what instructs our body to create the proteins it needs to run. Created by a four letter alphabet of ACGT, the human genome comprises a unique sequence that is over 3 billion letters long. These seemingly infinite combinations pass on genetic information from one generation to the next, and are fundamentally the reason we look like our parents. While the genome itself does not build us, it provides our cells with the information they need to do their jobs, such as maintaining our organs or responding to illness. In developments over the past years, the ACGT alphabet within DNA that builds and stores biological data has begun to be understood in relation to the codes seen within computing.
Despite formatting differences, organic data storage mechanisms such as DNA could be used to operate similarly to traditional digital data storage methods, such as the binary code. Over the past few years, developments show that the ones and zeros of binary code can be mapped to the ACGTs seen within DNA structures. This way of data storage and maintenance may seem far-fetched and unviable in current times, but this alternative method of data storage is being hailed as a substitute to traditional hard-drives, which are fast becoming environmentally and spatially unsustainable as the data produced in the world increases at an exponential rate. As outlined by AMPLYFI’s Dr. Lee Eccleshare in the whitepaper ‘Extracting Insight from Unstructured Data’, the amount of data we are producing is growing at an ever-increasing rate, with past estimates including a minimum of 2.5 quintillion bytes of data each day. Set in times of an incoming data storage crisis and increasingly accessible and cost-effective DNA sequencing, the level of motivation for recognising the potential for storing digital information within DNA has never been higher.
Every single day, DNA samples are being found and decoded from fossils that are hundreds of thousands of years old, showing that DNA is perfect for archival data storage over the long term. DNA data storage is fast becoming the most cutting edge solution to the long term issues expected with current mediums of data storage, such as corruption, degraded hardware and general matters of sustainability with regards to the manufacture of hundreds of millions of units of hard drives a year. DNA is now being seen as the ultimate upgrade from traditional hard-drives due to its ability to store data in a high-capacity, high-density and consistently available format, making it the gold standard of archival storage methods. However, DNA is by no means a perfect medium, largely due to its capability to mutate, which would be the organic equivalent of a corrupted harddrive. While there are coding mechanisms to mitigate the impacts of mutation on stored data, it progresses at a much slower rate than the encoding process itself. The science and technology behind DNA sequencing is falling into a similar pattern and is lagging far behind the standard of accuracy needed for a truly perfect retrieval of data stored within DNA.
Despite the DNA within a human genome weighing only 6.41 picograms (one trillionth of a gram), a single gram of DNA can theoretically store 215 million gigabytes of data, making it a small but extraordinarily mighty data solution. In scale this means that every piece of data recorded by the human race could be stored in the size and weight comparable to a couple of pickup trucks.
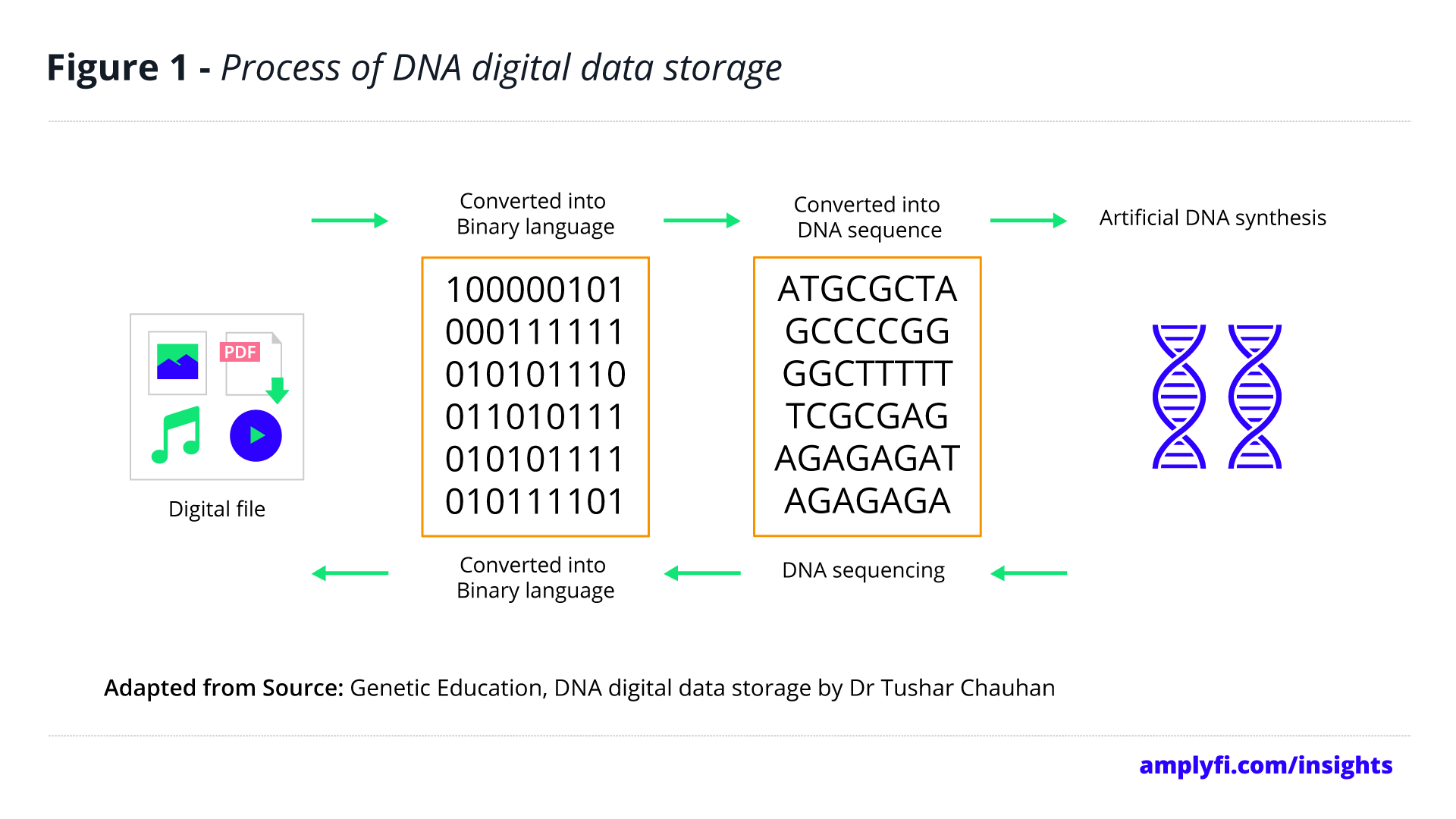
The process of DNA data storage can be summarised in three stages. The first stage being that of coding the data into the ACGT sequences seen in DNA, followed by linking the data into the DNA itself, otherwise known as synthesis. Following the logistic aspects of storage is the decoding process, where the DNA is sequenced to access the synthesised data. Due to the ability of DNA’s four-letter alphabet to be coded to and from binary, it has the potential to become a highly valuable resolution to common data storage problems. The four-component structure of DNA renders it able to store double the amount of traditional binary code.
In 2017, Erlich and Zielinksi were able to find that a ‘perfect retrieval’ of digital data from DNA is possible at a large scale, with 215 petabytes being able to be stored and decoded from per gram of DNA. Error-free decoding was enabled by the development of DNA Fountain, where the data is coded into DNA ‘Droplets’ which enable the data to be recovered even if there is a subset missing as lines of coded data are encoded into multiple ‘Droplets’. DNA Fountain demonstrates the potential for the domestic applications of DNA data storage, as Erlich and Zielinksi were able to not only encode a computer virus but a $50 Amazon gift card too, which was then decoded and then spent by an online follower of one of the developers. The possibility for the use of DNA data storage for domestic files was shown again by another group of researchers in 2020, where they were able to encode a file of Mozart’s sheet music and perform a perfect recovery of data from the encoded sample. The long term implications of the work of these researchers is yet to be seen in the public tech-sphere, but is steadily becoming the hottest topic within tech and academia spaces. Following Microsoft’s DNA Storage research project launch in 2015, it would come as no surprise if we were to see more household names enter the race to become the first to get large-scale DNA data storage into the hands of private sectors and the general public.
Originally an off-shoot from Oxford University, Oxford Nanopore Technologies is a company that has developed groundbreaking DNA sequencing technology that is not only portable but extremely affordable in relation to other more traditional means of sequencing, with some devices being available for just $1,000, with another device being able to sequence single samples for just $90. While this may still seem expensive, that initial $1,000 cost is entirely dwarfed by the cost of sequencing the first ‘draft’ of the human genome, which is estimated to have cost $300 million. The refining of that $300 million draft then cost a further $150 million, with the Human Genome Project acquiring $2.7 billion worth of funding despite finishing well over two years early, the $1,000 cost is seemingly affordable in comparison.
Ways of storing digital data need to be evolving just as quickly as the technology used to create it. In terms of technological developments, the environmental impacts of newer alternative modes of digital storage needs to be considered at both a consumer and corporate/institutional level, with discarded harddrives being viewed as having contributed to increased levels of toxicity at refuse sites due to the mixed and sometimes toxic metals they contain. DNA data storage is set to be among the latest developments in the technology field following the reams of published research in recent years hailing as a cure-all for the issues currently associated with traditional data storage methods. As the amount of data we create grows exponentially, we can expect to see more much needed developments in the data storage space.